STRUCTURAL GENOMICS
We perform theoretical and experimental studies to analyze and predict the 3D structure of the human genome.
Fields of research in lab
We perform theoretical and experimental studies to analyze and predict the 3D structure of the human genome.
We examine the relationship of the expression levels of selected genes from their location in 3D space.
To make sense of complex biological data, we apply machine learning techniques to extract multidimensional dependencies.
We use Deep Learning models for prediction of genetic features based on the sequence.
Bioinformatics services offered in our laboratory.
3DFlu is a comprehensive database designed to provide useful information regarding hemagglutinin proteins (HAs) of influenza virus type A.
The web-service that allows for creating 3D models of the genomic regions and their interactive visualization, as well as for a simple explorative data analysis.
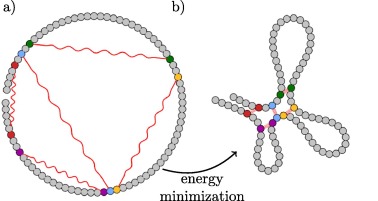
Spring model is a webservice for modelling chromatin domains using direct information about contacts derived from experiments like Hi-C or ChIA-PET. It is employs polymer physics and molecular mechanics using OpenMM library.
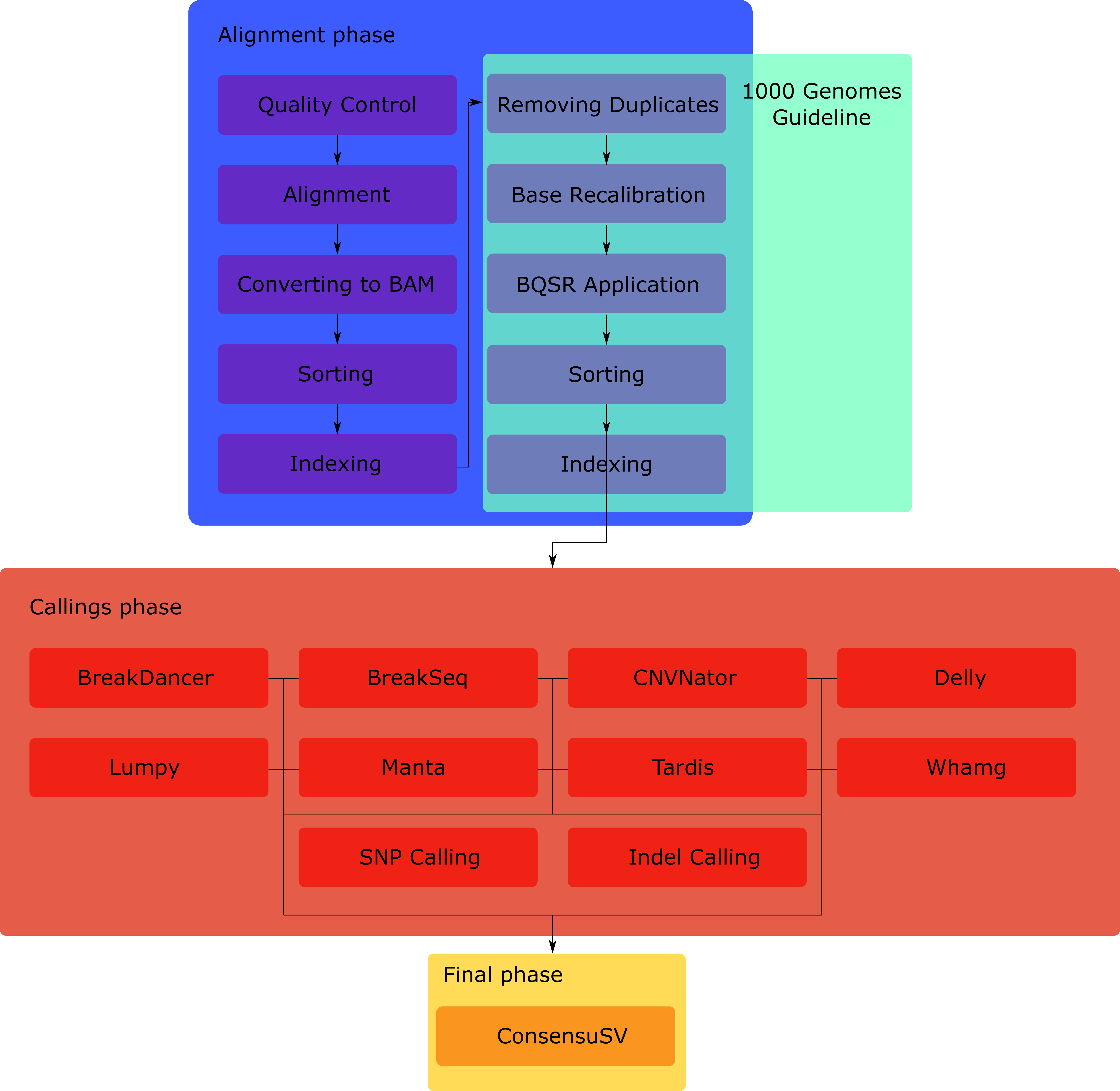
Automatised pipeline of ConsensuSV workflow, going from raw sequencing Illumina data (fastq), to output vcf files.
Three dimensional Genomics, 4DNucleome related projects, the higher order chromatin organization; Hi-C / ChIA-PET / Hi-ChIP experiments and data analysis, chromatin looping, topologically associating domains, compartmentalization of nucleus, lamina and subnuclear structures interactions with chromatin; genome wide association studies (GWAS), structural variants identification and functional analysis (including deletions, duplications, insertions, inversions, and translocations), next generation sequencing, short (Illumina) and long-reads (Oxford Nanopore, PacBio) DNA sequencing and data analysis, ChIP-seq data analysis from the 3D structure perspective; Big Data, statistical learning, massive dataset analysis, Computational Genomics and bioinformatics in population-level genomic data for medical applications and fundamental research in Life Sciences, structural and functional analysis of “omics” data; biophysical simulations , polymer modeling, DNA, RNA and proteins structure prediction, protein-protein, protein-DNA, protein-RNA and RNA-DNA interactions: prediction and analysis of biomolecular interactions networks; whole-cell systems biology modeling;
Project TEAM - Laboratory of Functional and Structural Genomics CeNT

Dariusz Plewczyński
Dariusz Plewczyński
Dariusz Plewczyński
Dariusz Plewczyński
Michał Łaźniewski
Kaustav Sengupta